The Fractured Entangled Representation Hypothesis
The Fractured Entangled Representation Hypothesis
Akarsh Kumar1,
Jeff Clune2,3,
Joel Lehman4,
Kenneth O. Stanley5
1MIT, 2University of British Columbia, 3Vector Institute,
4University of Oxford, 5Lila Sciences
Abstract
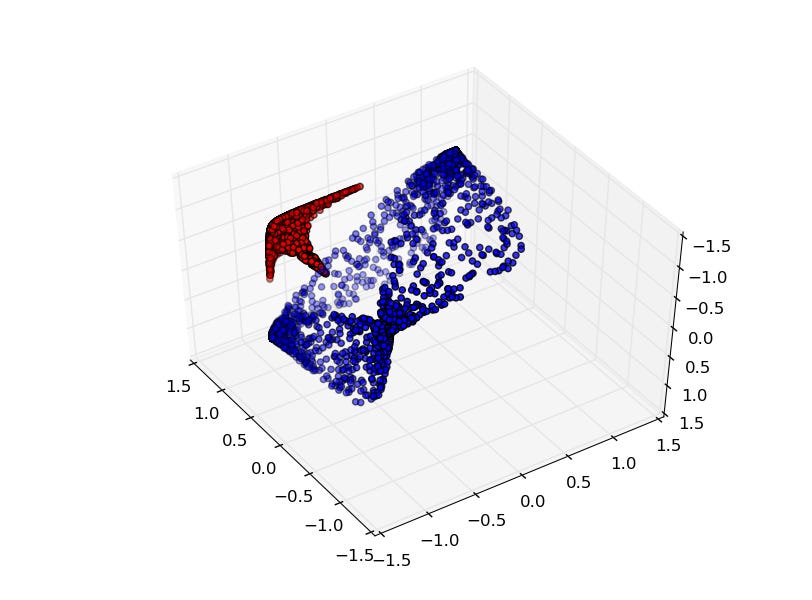
Much of the excitement in modern AI is driven by the observation that scaling up existing systems leads to better performance. But does better performance necessarily imply better internal representations? While the representational optimist assumes it must, this position paper challenges that view. We compare neural networks evolved through an open-ended search process to networks trained via conventional stochastic gradient descent (SGD) on the simple task of generating a single image. This minimal setup offers a unique advantage: each hidden neuron's full functional behavior can be easily visualized as an image, thus revealing how the network's output behavior is internally constructed neuron by neuron. The result is striking: while both networks produce the same output behavior, their internal representations differ dramatically. The SGD-trained networks exhibit a form of disorganization that we term fractured entangled representation (FER). Interestingly, the evolved networks largely lack FER, even approaching a unified factored representation (UFR). In large models, FER may be degrading core model capacities like generalization, creativity, and (continual) learning. Therefore, understanding and mitigating FER could be critical to the future of representation learning.
More Data and Visualizations




Here is the all of the supplementary data from the paper.
Intermediate Feature Maps:
- Skull:
- Butterfly:
- Apple:
All Weight Sweeps:
- Skull:
- Butterfly:
- Apple
Select Weight Sweeps From the Paper:
- Skull:
- Butterfly:
- Apple
Other Assets
All other important assets from the paper can be found in ./assets/.
Code
This repo contains code to:
- Load the picbreeder genomes from the paper
- Layerize it into a MLP format
- Train a SGD network to mimic that output
- Visualize the internal representation
- Do weight sweeps and visualize the result
Google Colab
For a quick start, open src/fer.ipynb in Google Colab:
Running Locally
To run this project locally, you can start by cloning this repo.
git clone https://github.com/akarshkumar0101/fer
Then, set up the python environment with conda:
conda create --name=fer python=3.10.16 --yes conda activate fer
Now you can install these libraries. The versioning needs to be very specific for the following. However, if you don't need to train the SGD network to play around with the results; in that case you can simply install the cpu version of jax following official guide.
python -m pip install jax==0.4.28 jaxlib==0.4.28+cuda12.cudnn89 -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html --no-cache-dir python -m pip install flax==0.10.2 evosax==0.1.6 orbax-checkpoint==0.11.0 optax==0.2.4 --no-deps
Then, install all the other necessary python libraries:
python -m pip install -r requirements.txt
Now you can run through the notebook src/fer.ipynb that quickly covers everything you need. However, here are some additional details on the file structure:
- ./assets contains the additional data and all assets from the paper
- ./picbreeder_genomes/ contains the raw picbreeder genomes for the skull, butterfly, and apple
- ./data/ contains the data for the layerized picbreeder CPPN and the SGD CPPN, we have already precomputed this directory for you
- ./src/ contains the code
- ./src/color.py is code to convert hsv to rgb
- ./src/cppn.py the flax CPPN code to model MLPs with arbitrary activation functions at different neurons
- ./src/process_pb.py processes the picbreeder genome to create layerized CPPNs
- ./src/train_sgd.py trains an SGD CPPN with a specified architecture on a target image
- ./src/util.py and ./src/picbreeder_util.py contain some utility functions
Contact
Please contact us at [email protected] if you would like access to more Picbreeder genomes for research.

Bibtex Citation
To cite our work, use the following:
@article{kumar2025fractured,
title = {Questioning Representational Optimism in Deep Learning: The Fractured Entangled Representation Hypothesis},
author = {Akarsh Kumar and Jeff Clune and Joel Lehman and Kenneth O. Stanley},
year = {2025},
url = {https://arxiv.org/abs/2505.11581}
}
What's Your Reaction?
 Like
0
Like
0
 Dislike
0
Dislike
0
 Love
0
Love
0
 Funny
0
Funny
0
 Angry
0
Angry
0
 Sad
0
Sad
0
 Wow
0
Wow
0